by Rami Major
Technique Name: Restriction Digest and Ligation
Fun Rating: 4/5
Difficulty Rating: 2/5
What is the general purpose? We use restriction digests to chew up DNA into smaller pieces, and ligation to stick them together.
Why do we use it?
If you want to use DNA like legos or puzzle pieces, mixing and matching different sequences together to create something new, restriction digest and ligation is for you!
What if you could be Dr. Frankenstein, but with DNA? Some of the DNA that we work with in the lab comes in the form of a plasmid. A plasmid is circular DNA that can be transformed into bacteria. As the bacteria grow, the plasmid DNA amplifies and can then be harvested from the bacteria. This DNA could contain the code to express a protein that you’re interested in, but sometimes, you might want to mix and match different proteins that you have, or combine multiple proteins into a single plasmid. This is where restriction digest and ligation come in. Restriction digestion breaks DNA up into different pieces, and ligation helps stick them back together!
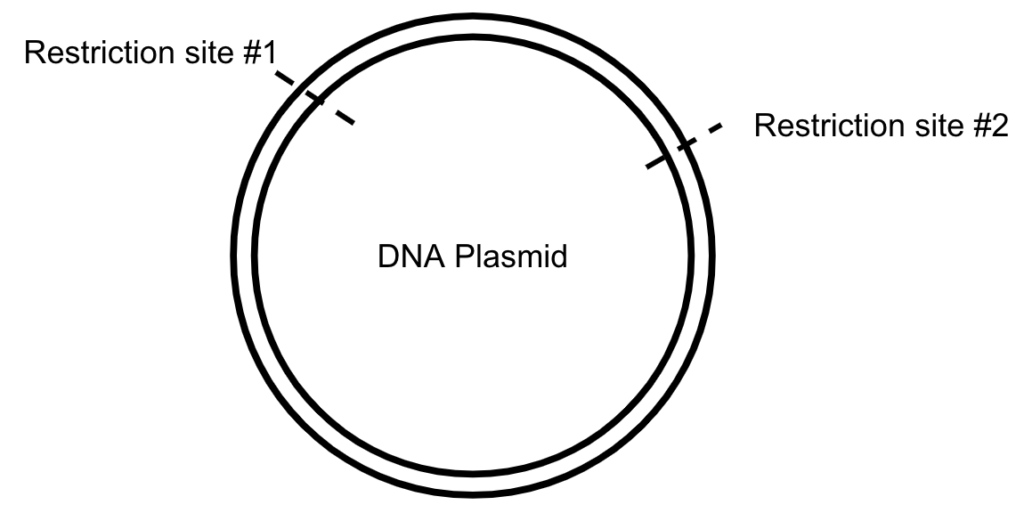
How does it work? Plasmid DNA contains sequences that are recognized by unique enzymes called restriction enzymes that cut DNA. These enzymes are “restricted” to only work at specific sequences of DNA. When restriction enzymes find their specific DNA sequence, they can cut the DNA, creating either “blunt ends” or “sticky ends.”
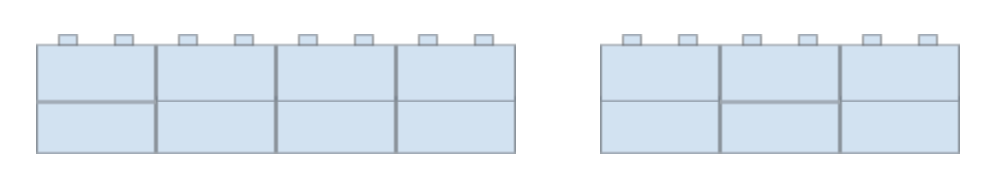
Blunt ends cannot be matched together with other sequences of DNA, as both DNA strands are cut in the same place. Picture legos: let’s say you’re building a stack of legos, two blocks high. You stack the legos directly on top of each other, and place these two-block high legos in a long line. The line isn’t connected, and you can easily break up that line by removing any of the legos and creating a blunt division (see image below).
Sticky ends on the other hand are created when the DNA is cut in different places on each strand, creating an overhang. This overhang could then match up with a new sequence of DNA, and you could stick these sequences together.

There are hundreds of different restriction enzymes, and each recognizes a unique sequence of DNA in order to make a cut. As a brief refresher, you might recall that DNA gets transcribed into RNA, which then gets translated into protein (following the central dogma of biology). So, Gene A would provide the DNA sequence needed to make Protein A. Here’s an example:
Goal: Produce Protein A in cells that Protein A is not normally expressed in
Idea: Tag Protein A with a fluorescent protein so that cells that produce Protein A will glow!
How to do this:
- Identify restriction sites that are located in both the plasmid that contains Gene A and the plasmid that contains the Fluorescent Gene. You want the restriction sites to be on either side of the Fluorescent Gene, so that you can cut it out of the plasmid!
- Use restriction enzymes to cut each plasmid, which will break up the DNA into smaller pieces.
- Run the DNA fragments on a gel using gel electrophoresis, which will allow you to separate the DNA pieces based on size.
- Cut the right bands out of the gel, and stick them in a tube together! This would mean you are putting together the DNA that has the Fluorescent Gene with the plasmid that contains Gene A.
- Add a special enzyme called ligase. Ligase uses the sticky ends created by the restriction digest and glues your pieces of DNA together.
- Voila! Now you have a plasmid that contains both Gene A and the Fluorescent Gene. You can add this to cells to express both Protein A and the Fluorescent Protein, and watch your cells glow!

The genes that create Protein A and a Fluorescent Protein are located in different plasmids, but you can use restriction digestion and ligation to put them in a single plasmid. When you add this new plasmid to cells, you can determine which cells are producing Protein A by noting which cells are glowing! (All figures and shapes created by the author in google docs.)
