by Tiffany Ko
Fun Rating: 3/5
Difficulty Rating: 4/5
What is the general purpose?
The Assay for Transposase-Accessible Chromatin (ATAC) using sequencing is used to find out which parts of a genome are open and available to be read. Similar to a recipe, DNA provides instructions on how to make the components of cells. When DNA is in an “open” state, the cell can read the DNA’s instructions.
Why do we use it?
DNA gives instructions on how to make components of a cell through a process called transcription. The instructions coded into the DNA, also known as genes, are copied by the cell’s machinery to make messages in the form of RNA. It’s crucial for your cells to be able to copy the DNA’s instructions into RNA messages that can travel to the protein-making parts of the cell.
Every cell in your body contains the same DNA, but your body is made of different kinds of cell types. Each cell type has its distinct shape and purpose: brain cells, heart cells, and skin cells all look and act differently from each other. This is because they are producing different kinds and different levels of proteins. How do the cells make certain proteins and not others?
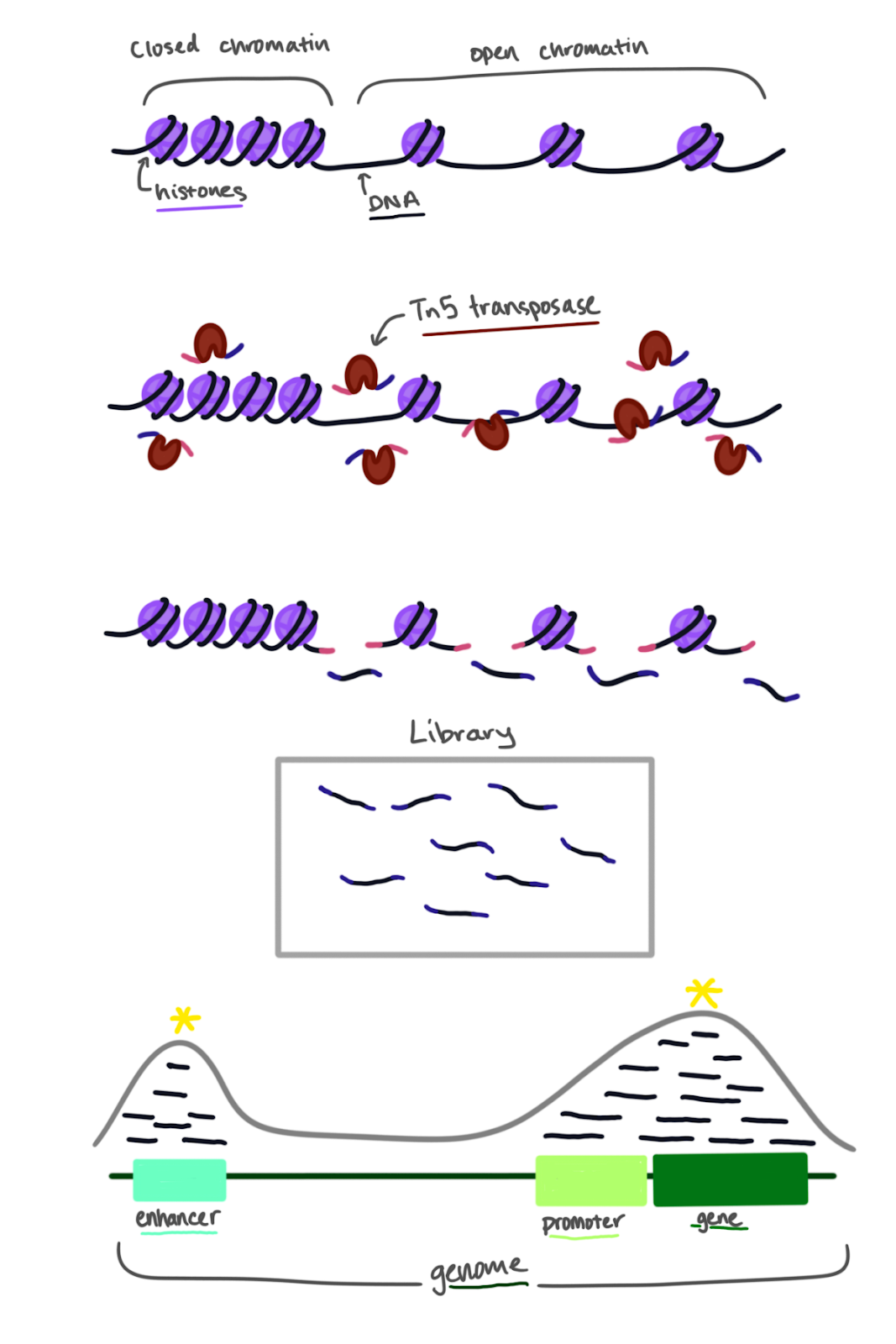
Although DNA is microscopically small, cells actually have a lot of it: for example, every human cell has an average of 6 feet of DNA inside it. Long pieces of DNA are folded and wound up around proteins called histones. These pieces of DNA wound around histones are known as chromatin. Sections of DNA can be more or less tightly packed, and this affects which genes are accessible to proteins that are critical for DNA transcription. The study of DNA packaging is called “epigenetics”. Epigenetics can change over time as you grow and develop, and it can also be influenced by the environment around you.
How DNA is packed can affect more than just the genes themselves. Right before the beginning of a gene’s DNA sequence, there is a short DNA sequence called a promoter that allows the cell’s machinery to copy the following gene into a message in the form of RNA. Besides genes and their promoters, there are also enhancers, which are sequences of DNA that enhance the cell’s RNA-making machinery to make copies of a certain gene. By using ATAC-seq, we can see if promoters and enhancers are in areas where DNA is packed more densely or more loosely.
How does it work?
- Cell Lysis (isolating the DNA)
For ATAC-seq, we need DNA from cells. The first step is to carefully lyse the cells, using a soap-like detergent to create holes in the cell membrane. This leads to the cells spilling out what is inside of them, including their nuclei (that contain their DNA) into the solution.
- Transposition (cutting DNA with a special enzyme)
What makes ATAC-seq unique is the use of a specially engineered enzyme called Tn5 transposase. The job of this particular enzyme is to cut DNA and then glue tags onto the resulting piece of DNA. Since DNA is packed tightly in some areas and loosely in others, the Tn5 transposase will only cut and tag the DNA in places that are less folded. You can imagine trying to cut a tightly wound ball of yarn compared to cutting a single strand of yarn. The Tn5 transposase snips the DNA in the loosely wound regions, which are more open and accessible. As the Tn5 transposase snips the open regions of DNA, it glues short tags onto the cut DNA pieces. These tags will be used in the next step of amplification.
- DNA Purification, Amplification, & Library Preparation
The cut and tagged DNA pieces have to be subsequently purified using special filters, and then amplified, or copied, multiple times through a process called the polymerase chain reaction.
- Next-Generation Sequencing and Analysis
The library made from the original cut and tagged DNA pieces is then sequenced using next-generation sequencing. The library is loaded onto a sequencing machine and each nucleotide base in the DNA is read. Once the pieces of DNA are sequenced, they can be lined up to the reference genome for the organism- humans, animals, plants, bacteria, and fungi all have reference genomes. Scientists can look for places in the genome where the cut and tagged pieces of DNA pile up (see figure). These peaks indicate that theTn5 transposase could snip the DNA at that location because it was open and accessible enough. The peaks can then be linked to genes, promoters, and enhancers. For example, in this illustration, we can see an example of a genetic region that has peaks of sequenced DNA aligned to the genome marked by a yellow asterisk. This means that at those locations (the enhancer, promoter, and gene in the illustration) the DNA was open and accessible for transcription.
ATAC-seq illustrated by Tiffany Ko
