by Anicka AbiChedid
Fun Rating: 3/5
Difficulty Rating: 3/5

What is the general purpose? Whole genome sequencing (WGS) is a technique used to identify the DNA sequence of a sample. This technique is especially useful for identifying any sequence changes, called mutations, that are present in the DNA.
Why do we use it? WGS is mainly used to identify a variety of mutations including single base substitutions, insertions and deletions of nucleotides, structural variants, and duplicates of chromosomes. Since WGS profiles the entire genome of a cell, it is often used clinically to identify variants responsible for causing cancer, which help tailor a patient’s treatment options. It is also useful for diagnosing genetic diseases and understanding population genetics.
How does it work?
Overview
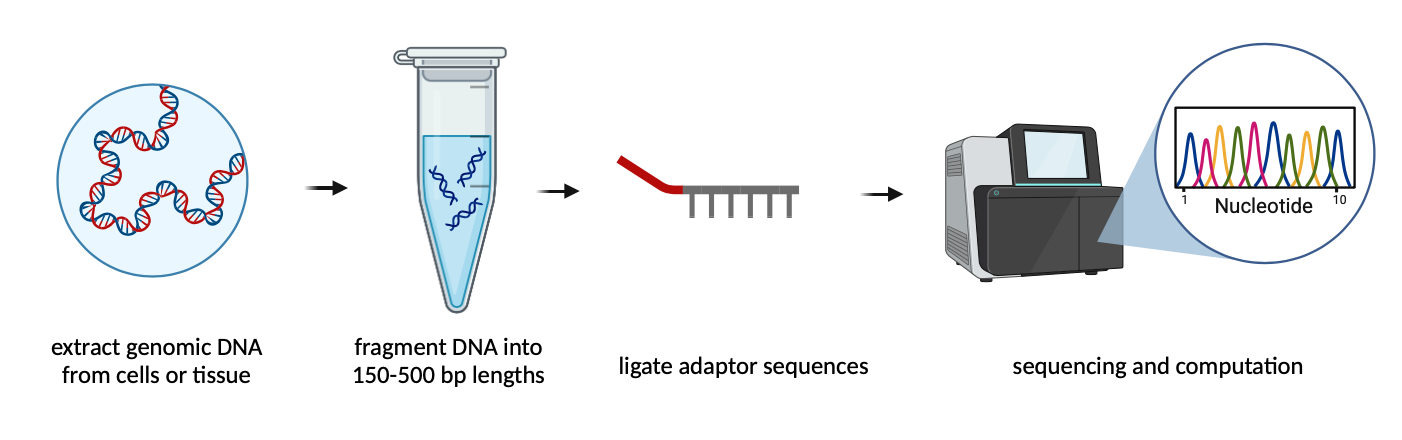
Figure 1. WGS is a multi-step process that involves fragmenting DNA into small pieces, attaching adaptor sequences to each fragment, binding the DNA to the sequencer, and computationally aligning and determining genomic variants. Image created in BioRender. Lab, S. (2025) ttps://BioRender.com/a92tnbr.
Steps
Library preparation
- DNA extraction: In order to access DNA inside the cell, we must break down cell membranes and proteins within the cell and leave the DNA intact. This can be accomplished using a lysis buffer, which disrupts the cell membrane, and a protease enzyme, which digests proteins. This process can be performed on many sample types, including cells and tissues, using commercially available kits.
- Fragment DNA: The extracted genomic DNA is then fragmented into smaller pieces around 150-500 base-pairs in length. This is performed using a sonicator, an instrument that uses high-frequency sound waves to break apart DNA.
- Ligate Adaptors: After the DNA is broken up into smaller pieces, another DNA sequence, called an adaptor, is added on to each piece. The adaptor contains sequences that identify the sample and bind the sequencing instrument. This step is performed by combining the fragmented genomic DNA and adaptors with ligase enzyme, which covalently links the adaptor to the DNA. This step can be performed using commercially available kits as well.
Sequencing
- Load Sample: The labelled DNA fragments are then loaded into the flow cell of a sequencing machine. The adaptor sequence on the labeled DNA binds to a complementary sequence in the flow cell.
- Perform Sequencing: After the DNA fragments are bound to the sequencer, a DNA polymerase amplifies the DNA using fluorescently labeled nucleotides. A camera captures the color of each nucleotide and repeats this process for every nucleotide on every DNA fragment. The sequencer then outputs a file containing the determined nucleotide sequences.
Data Processing
- Alignment: The output of the sequencer is called a FASTQ file, which contains the nucleotide sequences of the DNA fragments. These fragments are aligned back to the known genome using computational tools like BWA.
- Call variants: A second computational tool, such as GATK, determines any variants present in the DNA by comparing the nucleotide the sequencer called to the known reference sequence.
