by Caroline Aufgebauer
Fun Rating: 3/5
Difficulty Rating: 3/5
What is the general purpose? Ribonucleic acid (RNA) Immunoprecipitation Sequencing, or RIP-seq, is used to identify interactions between RNA and protein. It allows scientists to determine which RNA a protein binds to and where on the RNA it is bound.
Why do we use it? RNAs interact with many different proteins throughout their lifespan. These RNA-protein interactions help to regulate RNA in unique ways. For example, RNA-binding proteins (RBPs) can interact with RNA to transport it to specific locations within the cell. Proteins can also change the amount of RNA in the cell by stabilizing or degrading the RNA.
These RNA-protein interactions are highly dynamic. One way to think about this is how we change the clothing that we wear in response to temperature. A beanie “interacts” with our head to keep it warm. However, we place the beanie on and off our heads throughout the day in response to the ambient temperature. Likewise, proteins often have consistent binding sites, but their binding is highly dynamic in order to regulate RNAs in response to cellular signals. Thus, RIP-seq (Figure 1) is important because it allows scientists to not only identify RNA-protein interactions, but also understand how certain signals or disease states affect these interactions.
How does it work?
1. Cell Lysis
To access the RNAs and proteins within cells, we need to break the cells apart in a process called lysis. We lyse the cells using a detergent that destroys the cell membrane, releasing the inner cellular components. The resulting solution is transferred to a tube and referred to as cell lysate.
2. Immunoprecipitation
Next, we add antibodies that are attached to magnetic beads. These antibodies bind specifically to our protein of interest, allowing us to retrieve our protein and any RNA bound to it. When we attach our tubes to a magnetic rack, the beads along with our protein of interest are firmly attached to the wall of the tube by magnetic force. This allows us to remove other proteins and unbound RNAs through a series of wash steps (see Figure 1, Step 3).
3. RNA Isolation
At this point, we have successfully isolated the RNA-protein interactions that we are interested in. Now we need to separate the RNA from the protein so that we can identify each RNA. We add an enzyme called Proteinase K to break down proteins and release the RNAs from the magnetic beads. The RNA can now be purified through RNA extraction.
4. Library Preparation and Next-Generation Sequencing
We use next-generation sequencing to identify our RNA by reading the order of the four nucleotide letters (A, T, C, G) that make up each RNA. First, we convert the RNA into DNA to make it compatible for sequencing. Then, we add short sequences called adaptors to the ends of our DNA. These adaptors allow the sequencer to recognize the DNA. This process is referred to as library preparation. We can then run our library on a sequencer. The sequencer reads out the nucleotides in our DNA samples and gives us a list of the sequences.
5. Sequencing Analysis
We now have a list of the sequences that our protein binds to, but we do not know what genes these sequences belong to. We identify these genes by comparing our sequences to a reference genome. The reference genome is a known example of the complete sequences of genes of a particular species. We can map where our sequences are located in this reference genome. Proteins often have specific binding sites. Thus, the mapped sequencing reads often pile up around the same sites and generate peaks. We can then analyze these peaks to better understand what kinds of sites our protein of interest prefers to bind to.
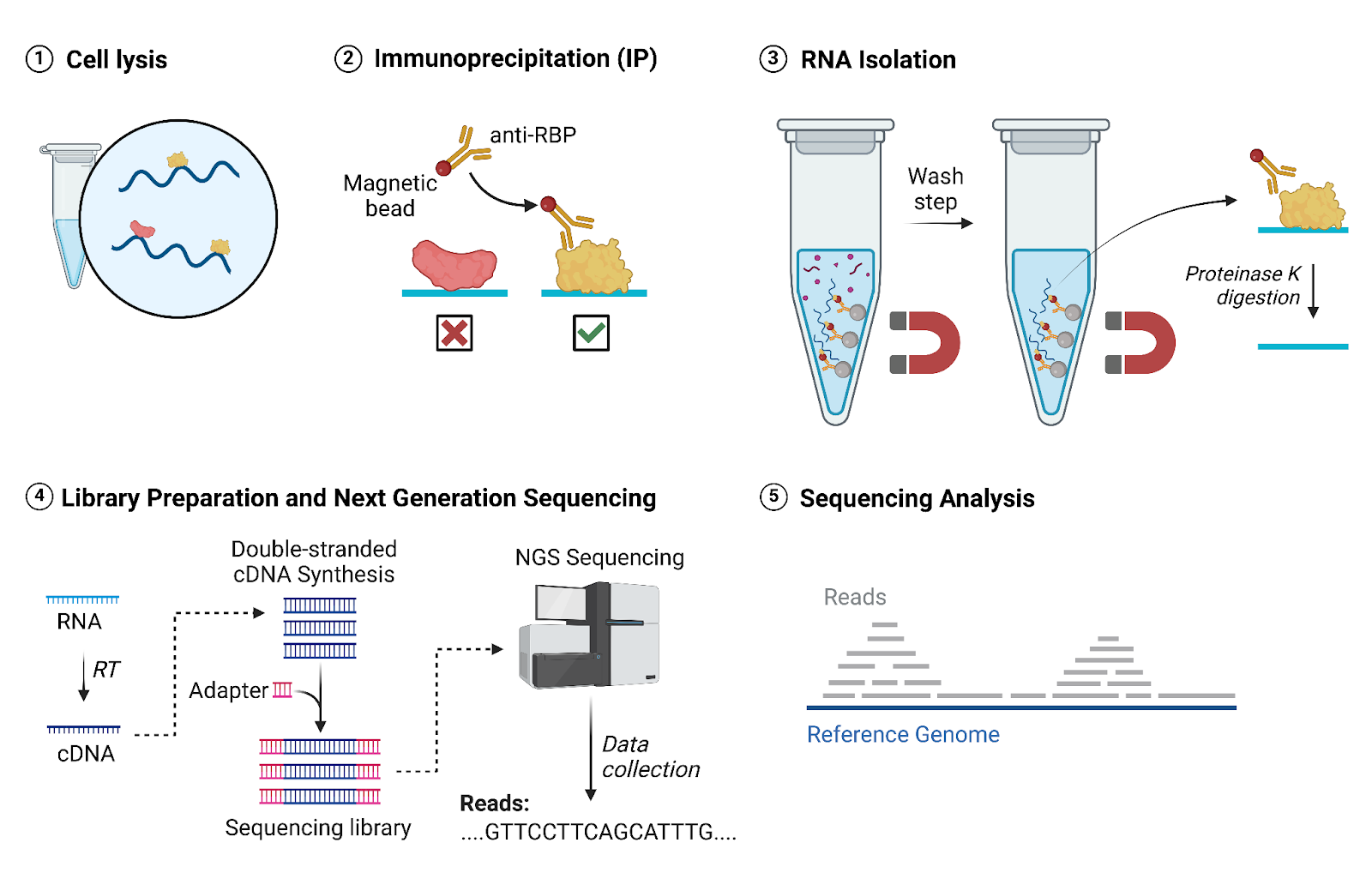
Figure 1: RIP-Seq Experimental Process. Image created by author using BioRender.
Conclusion: RIP-seq is a highly useful method that provides information about the binding patterns of protein on RNA. Knowing what RNA a protein binds to allows scientists to better understand how the protein regulates that RNA and ultimately cellular function.
