by Seth Veenbaas
Technique name: Sanger sequencing
Fun rating: 4/5
Difficulty Rating: 2/5

General purpose:
Sanger sequencing, and other DNA sequencing methods, are techniques used by biological scientists and medical doctors to read genetic messages within our cells. DNA sequencing identifies the exact order, or sequence, of the four unique nucleotide building blocks that make up our DNA (i.e. A, C, G, and T).
Why do we use it:
The information provided by DNA sequencing has long been a powerful tool for scientists to study genetics, evolution, molecular biology, and forensic science. Advances in DNA sequencing technology have also been used by medical professionals to diagnose diseases, identify specific variants of diseases and cancers, and even prescribe therapeutics personalized for patients.
History:
Sanger sequencing was developed in the 1970s by Fredric Sanger, who later won the 1980 Nobel Prize in Chemistry for this DNA sequencing technique. The Human Genome Project used Sanger sequencing to generate the first sequence of the 3 billion nucleotide-long human genome. This project was one of the largest scientific endeavors ever and took about $3 billion and 13 years to complete!
How does it work:
At its core, Sanger sequencing is a modified version of a technique called polymerase chain reaction or PCR. In PCR, a DNA strand called a template is copied several times to produce large quantities of DNA with identical sequences. The duplicate DNA strands are created by a DNA polymerase enzyme which reads the original DNA template and builds the duplicate strands one nucleotide (dNTP) building block at a time.
Sanger sequencing uses PCR with a special ingredient called a dideoxynucleotide (ddNTP). Usually, nucleotides (dNTP) have an oxygen atom that is used to link them together with the next nucleotide in a DNA strand, allowing the strand to grow in length. However, dideoxynucleotides are modified nucleotides that lack the oxygen atom needed to continue building the duplicate DNA strand. Because of this, using a small amount of dideoxynucleotides in a PCR reaction prematurely stops the reaction at a random location in the sequence. Randomly stopping the PCR reaction results in the creation of short DNA fragments of every possible length. Each type of dideoxynucleotide (i.e. A, C, G, T) is also labeled with a different color meaning that each DNA strand is colored based on the identity of the last nucleotide in the strand.
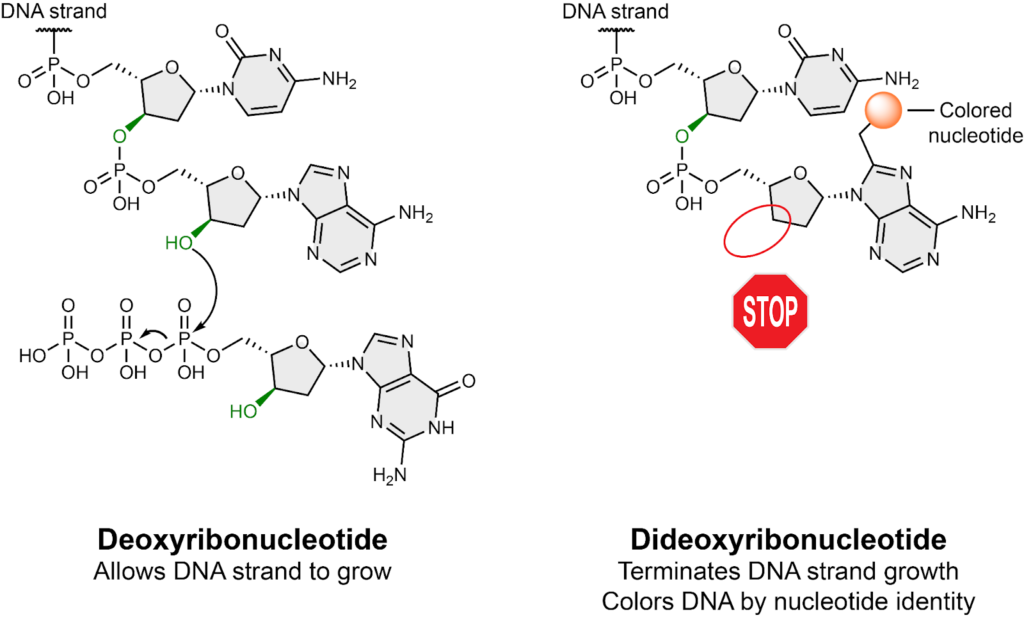
The last experimental step in Sanger sequencing is to separate all the colored DNA strands by their lengths. This is commonly done with either capillary electrophoresis or gel electrophoresis. (Fun fact: capillary electrophoresis was developed in North Carolina at UNC-Chapel Hill!) Lastly, the color of the sorted DNA strands is used to determine the identity of the nucleotide at every position in the sequence. Once all the nucleotides in the DNA are identified, the sequencing is complete!
Quiz: Sanger sequencing is limited to sequencing DNA with a maximum length of about 1000 nucleotides. How do you think the Human Genome Project was able to sequence a 3 billion nucleotide long genome with a method that can only sequence 1000 nucleotides at a time?
Answer: Long DNA strands must be cut into smaller fragments (< 1000 nucleotides) before they can be sequenced by Sanger sequencing. However, cutting the DNA into fragments creates a new problem because it is like cutting a complete picture into a bunch of shuffled-up jigsaw puzzle pieces. Similarly to how a jigsaw puzzle is solved, sequences from each DNA fragment need to be combined together in the correct order (or aligned) to solve the full-length DNA sequence. Luckily, computer alignment software now exists to solve this problem and determine the full-length sequence.

Conclusion: Sanger sequencing is a highly accurate (~ 99.99%) sequencing technique that is frequently used by scientists in addition to other complementary sequencing methods. Together, DNA sequencing techniques facilitate modern biological science, modern medicine, and even at-home ancestry DNA testing kit.
If you want to try DNA sequencing at home read our article on commercial DNA testing kits.
