by Olivia Conway
Fun Rating: 3/5
Difficulty Rating: 2/5

What is the general purpose?
Although the genetic engineering we read about in science fiction novels is still far in the future, molecular biologists use proteins and chemicals to edit DNA every day! One of the easiest ways to do this, known as the Gibson Assembly, was developed by Dr. Daniel Gibson in 2009 as an improved method for incorporating multiple pieces of DNA into a single circular nucleic acid structure. The ability to quickly generate specific DNA sequences has revolutionized the fields of molecular and synthetic biology. For example, the pharmaceutical industry has shifted towards developing targeted molecular therapies that use biological enzymes to treat illnesses like cancer. The Gibson Assembly procedure simplifies the assembly of several different enzymes into a single DNA sequence and allows scientists to swap out proteins to produce several different drugs based on the same structure. Additionally, Gibson Assembly can be used to insert additional genes into bacterial or yeast genomes, which provides new opportunities for engineering bacteria that can contribute to energy production or waste removal.
Why do we use it?
Many molecular biology experiments require that an organism produces edited genetic material, like a specifically mutated DNA sequence or a gene tagged with green fluorescent protein (GFP). These sequences are inserted into a circular nucleic acid sequence called a plasmid, and the plasmid is introduced to cultured cells so the gene or mutation can be studied. Most labs have an extensive collection of plasmids with different fluorescent tags, promoters, and genes for antibiotic resistance, but sometimes scientists need to move a gene from one plasmid to another. Previous techniques relied on restriction enzymes, which are proteins that cleave specific sequences of DNA so that sections of DNA can be pasted together. This method can be quite complicated because it relies on multiple restriction enzymes that need different buffers and incubation temperatures. In contrast, the Gibson Assembly uses a trio of enzymes that work at the same temperature to rearrange multiple pieces of DNA into a single circular sequence in only one step.
Additionally, Gibson Assembly is so powerful that scientists have used it to assemble entire small genomes from hundreds of short genetic sequences. The research group that pioneered the Gibson Assembly assembled a 583 kilobase bacterial genome with their new technique, and it has since been used to assemble yeast genomes and mammalian mitochondrial genomes.
How does it work?
Part 1: Amplifying Desired Sequences
Primers are single-stranded DNA sequences that can bind to a gene of interest for amplification during polymerase chain reactions (PCR). Before a Gibson Assembly can be performed, the genetic sequences that are going to be joined together must be isolated and amplified. Using the example in Figure 1, the scientist would need primers to amplify their gene of interest and primers to amplify the red fluorescent protein (RFP) plasmid (Figure 1). A Gibson Assembly only works if the sequences overlap, so the primers for the gene of interest must include a sequence that overlaps with the plasmid.
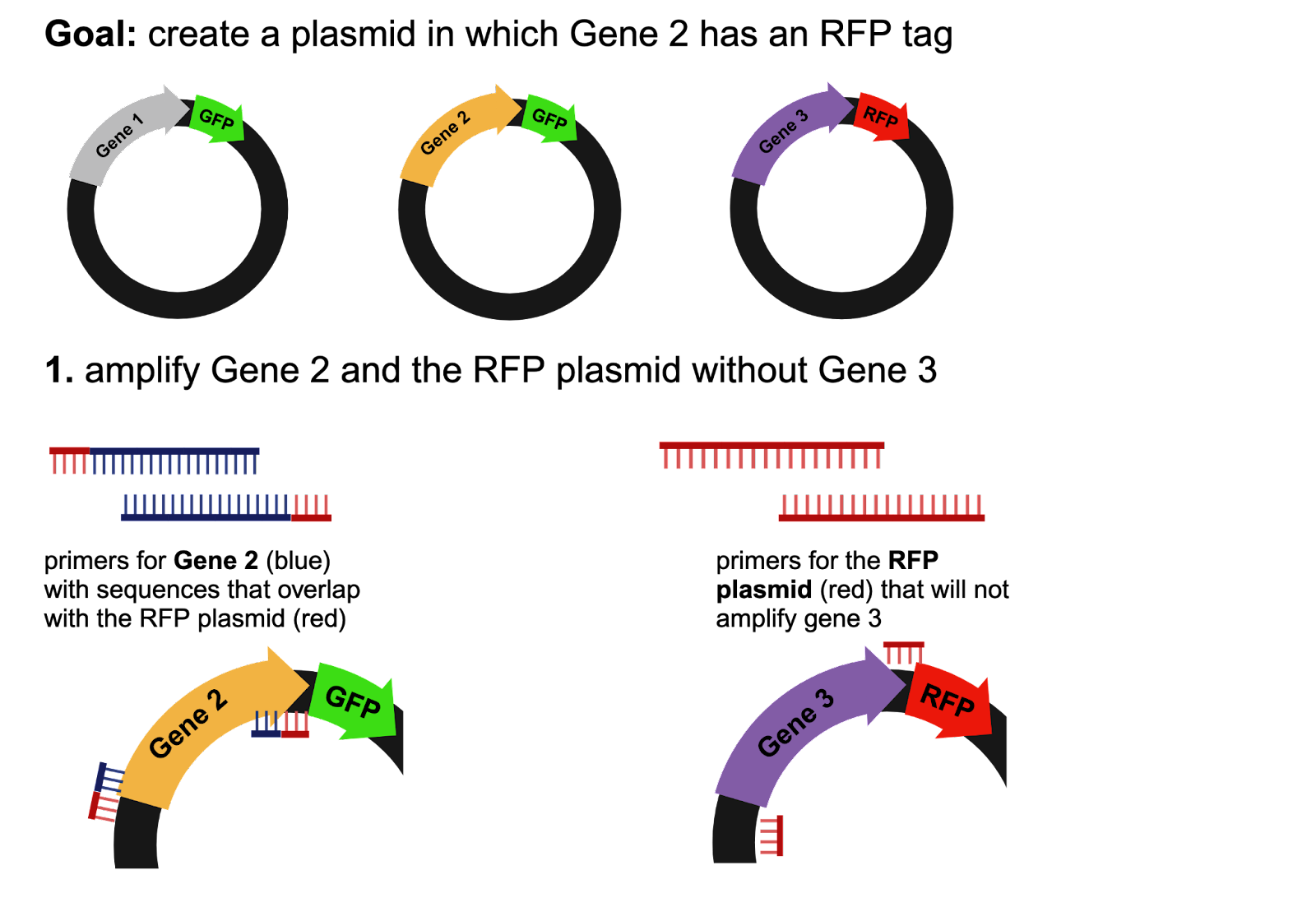
Figure 1
Part 2: Gibson Assembly
Once the DNA for the experiment has been obtained, the Gibson Assembly itself is quite simple. To assemble the new plasmid, the DNA sequences are mixed in a tube with three proteins (5’ exonuclease, DNA polymerase, and DNA ligase) and incubated at 50°C for 15 minutes (Figure 2). This incubation step can be done in a PCR thermal cycler or a standard incubator.
During the incubation, the 5’ exonuclease removes the 5’ end of each DNA sequence to generate “sticky ends”. These “sticky ends” can then be fused together due to the overlap between the original sequences. Once the complementary sequences have been stuck together, the DNA polymerase synthesizes new DNA nucleotides to replace the sequence that the 5’ exonuclease destroyed. Finally, the DNA ligase forms a bond between the new sequence and the original sequence to seal the DNA strand.
After the incubation, the new plasmid can be used for downstream procedures like bacterial transformation and protein purification.
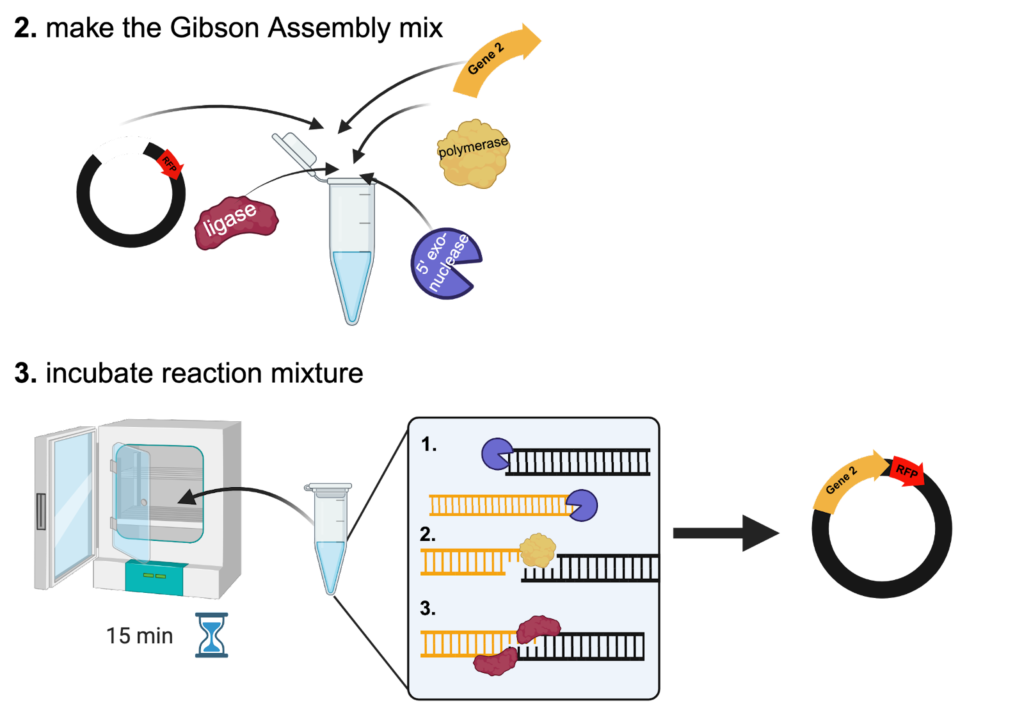
Figure 2
