by Hazel Milla
Fun Rating: 4/5

Difficulty Rating: 4/5

What is the general purpose?
Mass cytometry, also called cytometry by time-of-flight (CyTOF), is a technique that allows scientists to identify the types of cells in a sample, such as blood or tissue. It also allows us to characterize how cells respond to various stimuli. For example, CyTOF has been used to identify the immune cells, or the cells that protect your body against disease, that are present in the blood during infection with COVID-19. Characterizing immune cells during an illness can tell us how the immune system responds to the infection.
Why do we use it?
The immune system faces a host of potential problems: damaged tissue that needs repair, cancerous cells that need to be repressed, and infection caused by bacteria, viruses, fungi, or parasites. To address these issues, the immune system has many cell types which carry out appropriate, effective responses against harmful stimuli. This response consists of recruiting specific cells to sites of infection or damage. In these locations, the immune cells prompt useful cells to divide and injured cells to self-destruct. Immune cells can also be activated. Activation can mean many things in this context, from producing proteins that recruit other cell types to killing infected cells. Whether or not a cell divides, dies, or activates in a specific way depends on its type and function.
We can use CyTOF to determine which cells are set in motion and activated by the immune system in response to harmful stimuli. This allows us to better understand how our immune system works and what might happen when things go wrong (for example, when the immune system cannot fight off an infection like COVID). Furthering our understanding of the immune system in this way can help us develop treatments and diagnose diseases. For example, scientists have been able to identify different immune cell compositions in people who develop long COVID after infection vs those who do not. This knowledge could aid in developing preventative measures and treatments for long COVID.
If you’ve read about flow cytometry, you may notice that CyTOF and flow cytometry are used for similar reasons. With both methods, scientists can characterize cell types to develop treatments and diagnose disease. This prompts the question: Why not use flow cytometry? Unlike flow cytometry, CyTOF allows us to characterize cell types more extensively while requiring less blood or tissue. So, while flow cytometry is incredibly useful, sometimes it is preferable to use CyTOF.
How does it work?
CyTOF integrates techniques from mass spectrometry and flow cytometry to identify cell types. To give you a better understanding of CyTOF, let’s briefly summarize the two methods it’s based on:
Flow cytometry: Flow cytometry connects cellular proteins with fluorescently tagged antibodies (proteins produced by the immune system used to recognize substances). These cellular proteins can be on the outside of cells, or scientists can use chemicals to poke holes in the cells to tag the proteins that are located inside of cells. When exposed to lasers, these fluorescent proteins emit light at specific frequencies.
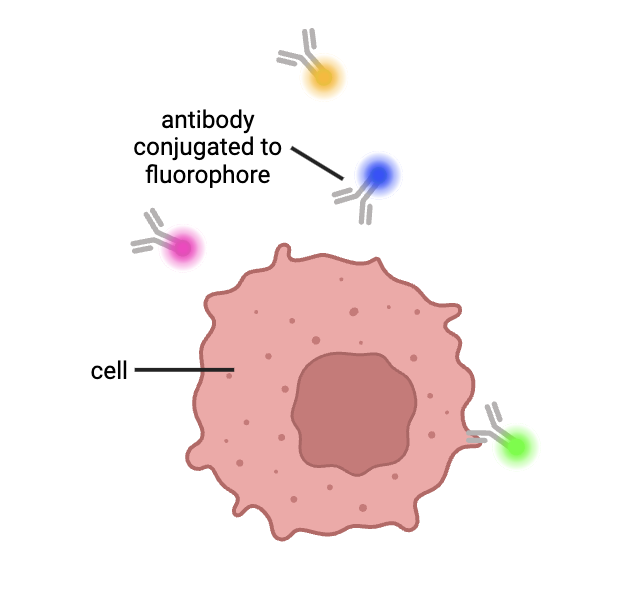
Figure 1: Cells are labeled with antibodies attached to fluorescent molecules. These antibodies bind to specific proteins that a cell expresses. Figure created by author using BioRender.
After being tagged with fluorescent proteins, cells are moved through a machine, which directs lasers at them. A detector then picks up the signal emitted by the tagged cells, which allows us to identify the proteins they express and thus the types of cells in the sample.
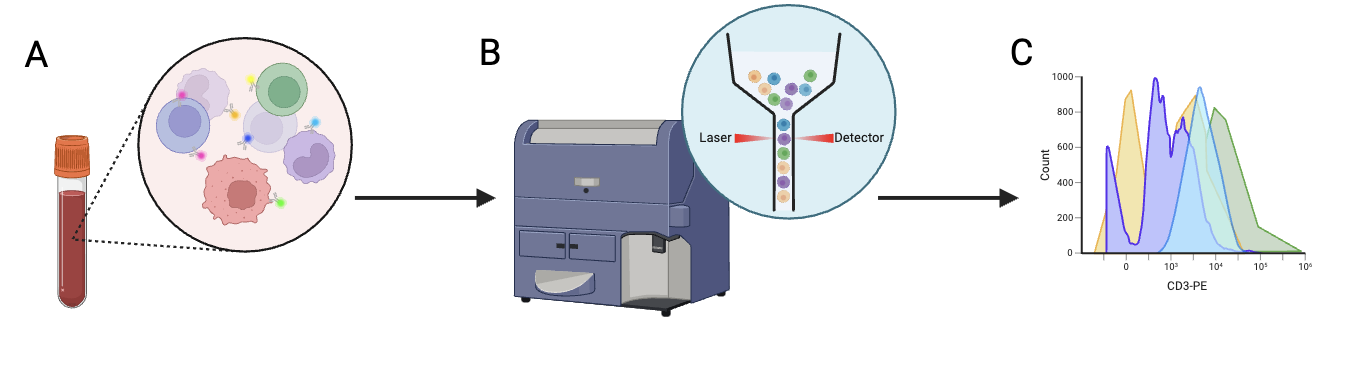
Figure 2: A) Cells are labeled with fluorescent proteins. B) Cells are placed in a machine that organizes the cells into a line before they are run through the laser one by one. Their signal is picked up by a detector on the opposite side of the laser. C) The signal detected by the machine is used to identify the cell type. Figure created by author using BioRender.
Mass spectrometry: Mass spectrometry characterizes molecules based on their molecular weight. To begin the process, molecules are isolated using a separation technique called chromatography. After the molecules are isolated, the sample is injected into a machine, where it is vaporized (converted to gas) with heat. Electrons are added to the molecules in a process called “ionization.” Through this process, each molecule develops a charge because its number of electrons has changed (and is therefore imbalanced). After vaporization and ionization, the molecules are exposed to a magnetic or electric field. The field manipulates the ions, or charged particles, so that they move at different rates based on their size and charge. A detector then determines how many ions reach it as a function of time. From this, we can identify the ions’ mass-to-charge ratios, enabling us to work out the structure of the molecules in our sample.
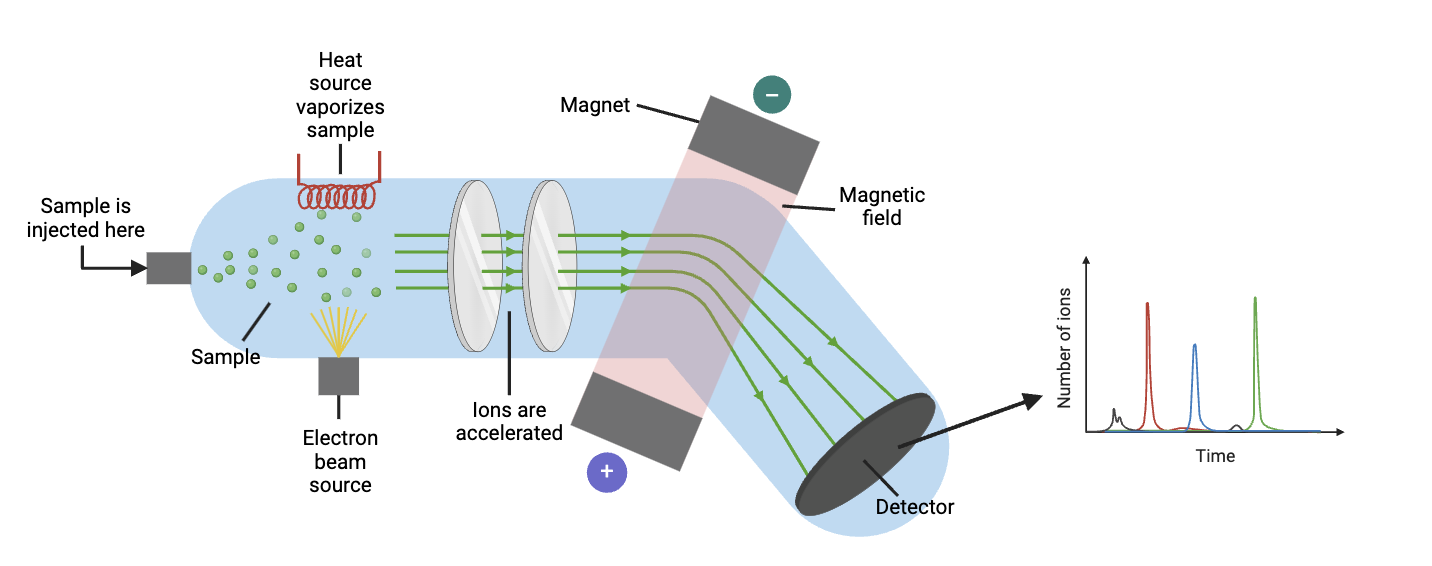
Figure 3. Overview of mass spectroscopy. Figure created by author using BioRender.
How these methods relate to CyTOF: Like flow cytometry, CyTOF allows scientists to identify cell types by tagging their proteins with antibodies. With CyTOF, however, antibodies are connected to heavy metal ions instead of fluorescent proteins. Using heavy metal ions helps us avoid a setback in flow cytometry: overlapping fluorescent signals. When there are a lot of proteins we want to look at, we need to use more fluorescent proteins, which translates to even more overlap, making it harder to differentiate signals. By getting around this limitation of flow cytometry, CyTOF allows us to tag more proteins than with flow cytometry. This allows us to gain more information about our cells with fewer assays, which also means we can use CyTOF when blood or tissue samples are limited.
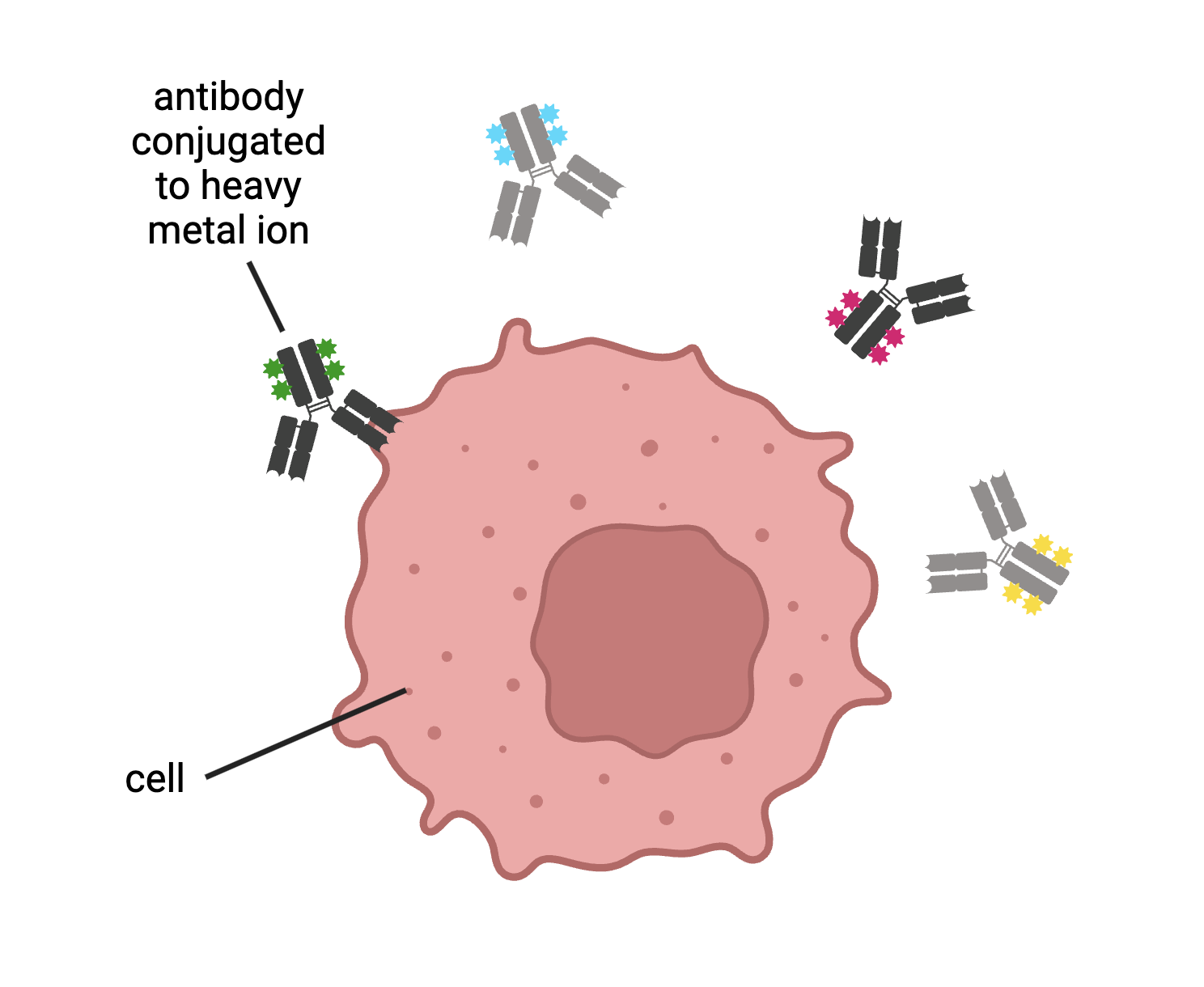
Figure 5. With CyTOF, antibodies are conjugated to heavy metal ions. These antibodies then bind to proteins expressed by cells. Figure created by author in BioRender.
After tagging our proteins, we vaporize and ionize our sample (much like with mass spectrometry), turning it into a cloud of ions. This cloud of ions is sent through a time-of-flight analyzer. Each ion’s charge and size are identified based on the time it takes the ion to reach the detector (in other words, its “time of flight”). From this data, we can figure out which antibodies, and thus which proteins, were present in our original sample. Knowing the proteins in our sample can help us determine which cell types were present. It also gives us insight into what the cells were doing when tagged (for example, if they were dividing or producing activating proteins).
To summarize, CyTOF is a method that takes advantage of flow cytometry and mass spectrometry principles to increase the number of proteins that can be tagged in a sample. By increasing the number of tagged proteins, we can characterize the cells in our sample with greater depth and precision. This helps clinicians and scientists diagnose disease and develop treatments more efficiently and effectively. With COVID-19 in particular, CyTOF has been a useful tool in characterizing immune responses associated with severe disease and long COVID.
