by Kerstin Baran
Fun Rating: 3/5

Difficulty Rating: 5/5

What is the general purpose? Spatial transcriptomics is a cutting-edge technique that combines the spatial (location) data of intact tissues with the gene expression (transcription) data from RNA sequencing experiments. The main question that spatial transcriptomics experiments are used to answer is: What is the transcriptome of cells in a location of interest in a tissue section?
Why do we use it? Spatial transcriptomics is a powerful technique because it enables researchers to know what genes are being expressed in specific regions of tissue. Traditional RNA sequencing experiments lose spatial information because you have to mix the tissue of interest in a tube, destroying the structural integrity of the tissue. In spatial transcriptomics, the RNA from cells is extracted from a thin slice of an organ (tissue) on a slide, and the RNA can be traced back to certain cells or areas of the slide.
How does it work?
Spatial transcriptomics experiments require specialized machines that can both capture an image of the slide and collect RNA directly from the cells on the slide without disrupting the tissue. Multiple companies make platforms to perform spatial transcriptomics experiments, and they each use different proprietary chemistry to extract the RNA. However, the main steps and concepts are the same.
First, a researcher will prepare a tissue of interest for histology, which includes fixing the tissue, embedding it in paraffin wax (for preservation of the organ structure), and slicing a very thin section of the tissue onto a slide. In most cases, the slide can be stained for cell-type specific markers using immunofluorescence, which can help determine what region of the tissue is of interest to the researcher. Spatial transcriptomics can be done with human and mouse tissue sections, giving researchers the option to use tissue from mouse models and from clinical/patient samples.
Next, in most cases, RNA probes with spatial markers are applied to the tissue on the slide. These probes hybridize, or “stick to” the RNA in the tissue. The spatial marker on the RNA will allow the RNA to be mapped back to a specific location on the slide. From there, the machine will capture an image of the slide and the RNA will be sequenced.
Finally, the sequencing data is processed and made ready for the researcher to analyze. Spatial transcriptomics empowers researchers to capture the gene expression from specific areas of tissues. For example, a cancer researcher can now compare the gene expression between the cells immediately surrounding a tumor (part of the tumor microenvironment), cells distant from the tumor, and the tumor itself. This cutting-edge method is revolutionizing science and has even won Nature’s Method of the Year in 2020.
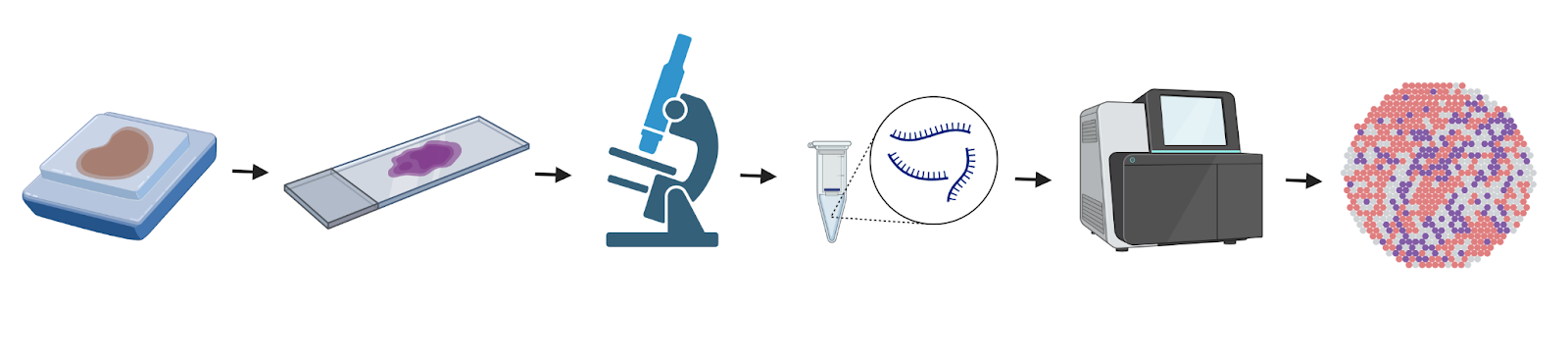
Image 1. Spatial transcriptomics workflow: processing tissue sample, sectioning sample onto slide, image capture, RNA extraction, RNA sequencing, and analysis. Created with BioRender.com
